Researchers Propose Identifying Strategies for Microbial Utilization of Exogenous Substrates Based on Molecular Biological Techniques
Soil microorganisms act as executor in mediating soil organic carbon (SOC) turnover. The huge soil microbial pool harbors complex microbial community compositions, how and to what extent microorganisms regulate SOC turnover critically depend on their substrate utilization strategies.
Whereas, how microbial substrate utilization strategy regulate such non-specific processes in carbon (C) sequestration still remain unclear in science. “To know ‘who does what’ is a prerequisite when linking the functional traits of different microbial taxa in substrate transformation in soil,” pointed by Prof. ZHANG Xudong and Prof. HE Hongbo from the Institute of Applied Ecology of the Chinese Academy of Sciences.
In their recently published study entitled “Identification of microbial strategies for labile substrate utilization at phylogenetic classification using a microcosm approach” in Soil Biology and Biochemistry. In this research, they conducted a simulated microcosm amendment with weekly addition of 13C-glucose and microbial utilization of the extraneous C was identified with the approaches of nucleic acid stable isotope probing (DNA-SIP) and high-throughput pyrosequencing techniques.
Despite of the increasing C availability with glucose addition, the phyla of Proteobacteria and Actinobacteria retained the preference on glucose while the growth of Acidobacteria and Chloroflexi was dominantly dependent on native SOC, suggesting that the substrate preference of bacteria was intrinsically controlled by their copiotrophic or oligotrophic patterns. Such substrate utilization strategies of bacteria at phylum level were primarily originated from the genus level with phylogenetically conserved attributes. Comparatively, the dominant phyla of Ascomycota, Basidiomycota and the dominant genera of fungi could utilize both labile C and native SOC, indicating their wide spectrum of substrate utilization strategies.
Based on these results, they found the distinct substrate utilization strategies of bacteria and fungi and thus exhibited a clear picture of ‘who was activated to eat labile C’. Such findings could link the functional traits of microorganisms to their distinct roles in regulating SOC cycling,” which have important implications in the elucidation of their functional traits involved in SOC turnover.
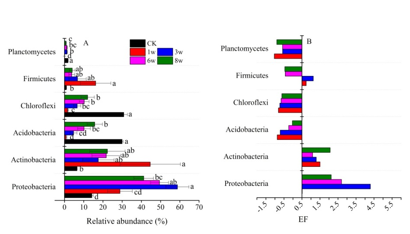
Fig. 1 Relative abundance (means ± SE) of dominant bacterial phyla in 13C-DNA (A) and the enrichment factors (EF) of dominant fungal phyla in 13C-glucose treatments (B) (Image by WANG Xinxin).

Fig. 2 Relative abundance (means ± SE) of dominant fungal phyla in 13C-DNA (A) and the enrichment factors (EF) of dominant fungal phyla in 13C-glucose treatments (B) (Image by WANG Xinxin).
This study was supported by The Natural Science Foundation of China, National Key Research & Development Program and the “China Soil Microbiome Initiative: Function and regulation of soil - microbial systems” of the Chinese Academy of Sciences.
Publication Name: ZHANG Xudong et al.
Email: yueqian@iae.ac.cn



