The Discovery, Identification, and Naming of New Species of Organohalide-respiring Bacteria
Dehalogenimonas, a genus within the phylum Chloroflexota, belongs to the class Dehalococcoidia. Its members are strict anaerobic, obligate organohalide-respiring bacteria that derive energy through reductive dehalogenation reactions mediated by reductive dehalogenases. Previously identified species of Dehalogenimonas can only use halogenated alkanes as electron acceptors and carry out organohalide respiration through dichloroelimination reactions. Currently, there is a limited number of pure cultures of Dehalogenimonas available. The discovery and isolation of new strains of Dehalogenimonas are crucial for expanding the biodiversity within this genus, enhancing our understanding of their ecological functions, and exploring their potential applications in pollution management.
The research team at the Institute of Applied Ecology, Chinese Academy of Sciences, focusing on "Microbial Ecology of Contaminated Environments," collaborated with the University of Tennessee, USA. Their joint efforts led to the discovery and enrichment of a novel Dehalogenimonas strain capable of utilizing various chlorinated alkenes, including vinyl chloride, for growth. This strain was isolated from grape pomace compost samples. Through advanced techniques such as metagenomics and metaproteomics, the gene responsible for vinyl chloride reductive dehalogenase was identified, leading to the naming of the strain as 'Candidatus Dehalogenimonas etheniformans' strain GP. Previous studies had only identified certain strains of Dehalococcoides capable of respiratory metabolism for dechlorination of vinyl chloride, a known human carcinogen. The discovery of the GP strain expands the microbial resources available for remediating environments contaminated with organochlorine compounds.
The research team successfully isolated strain GP under strict anaerobic culture conditions and conducted a comprehensive classification and identification study using multiple methods, including whole-genome sequencing, comparative genomics analysis, phenotypic characterization, and physicochemical property analysis. The goal was to gain a comprehensive understanding of the evolutionary status, physiological and biochemical characteristics, and environmental remediation potential of strain GP. Compared to other known organohalide-respiring bacteria, the genome of strain GP contains the highest number of homologous genes encoding reductive dehalogenases. Phylogenetic analysis based on the 16S rRNA gene and genomic sequences supports the conclusion that strain GP represents a new species within the genus Dehalogenimonas.
Physicochemical identification experiments revealed that strain GP shares certain similarities with reported strains of the genus Dehalogenimonas in terms of phenotypic characteristics, phospholipid fatty acid composition, and optimal growth conditions. However, it exhibits distinct differences in electron acceptor utilization. Strain GP efficiently dechlorinates various chlorinated ethenes, including vinyl chloride, and 1,2-dichloroethane via hydrogenolysis, resulting in the complete dechlorination to non-toxic ethene. Through collaboration with Professor L?ffler's team at the University of Tennessee, USA, we classified strain GP as a new species within the genus Dehalogenimonas and formally named it Dehalogenimonas etheniformans.
This study has further improved our understanding of the physiological and evolutionary characteristics of Dehalogenimonas members, providing theoretical support and strain resources for the bioremediation of halogenated hydrocarbon-contaminated sites. Strain GP has been deposited in the China General Microbiological Culture Collection Center (CGMCC) and the Biological Resource Center of the National Institute of Advanced Industrial Science and Technology, Japan (JCM).
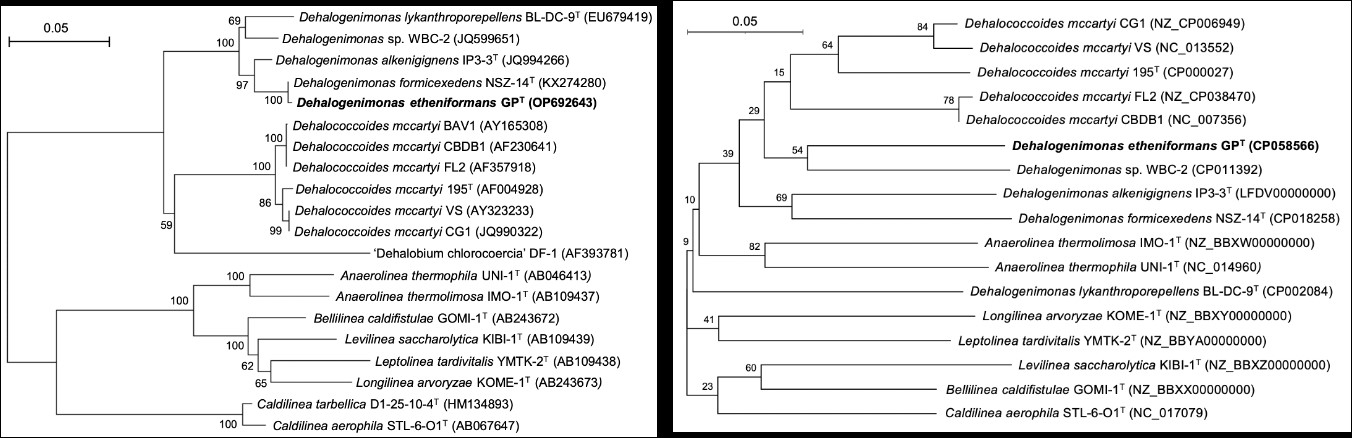
Fig.1 The phylogenetic tree analysis of strain GP based on the 16S rRNA gene sequences and whole-genome sequences (Image by YANG Yi).
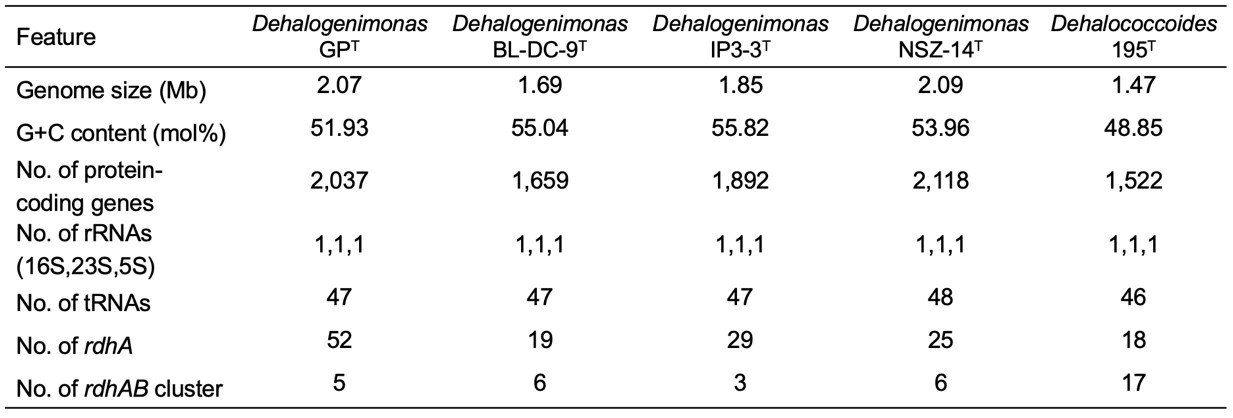
Tab.1 Comparisons of the genomic features of strain GP with representative strains from the genera Dehalogenimonas and Dehalococcoides (Image by YANG Yi).
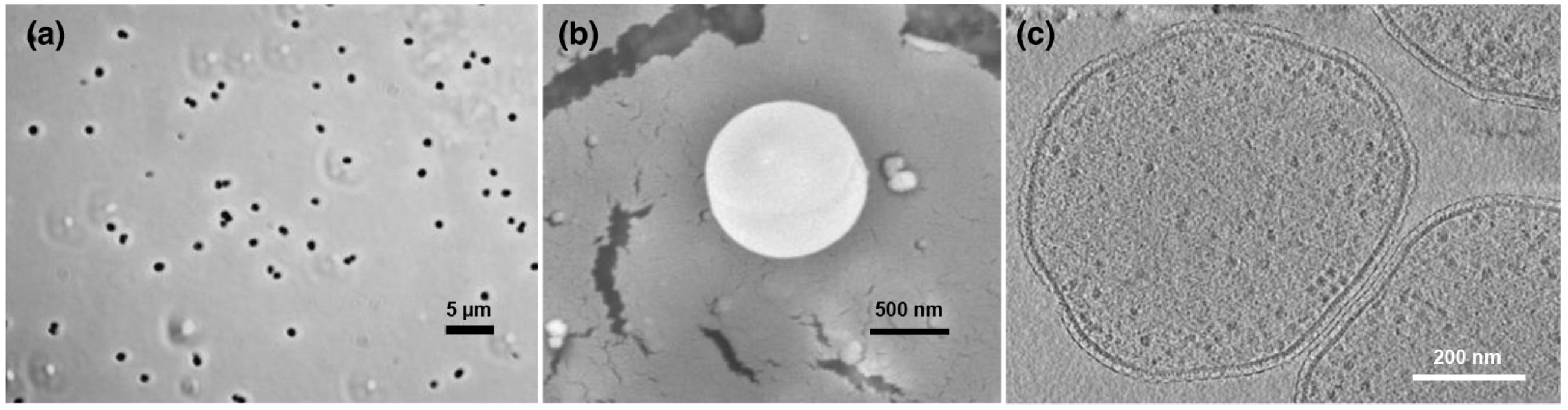
Fig. 2 The differential interference contrast (DIC) microscopy, scanning electron microscopy (SEM), and cryo-electron tomography (Cryo-ET) high-resolution images of strain GP (Image by YANG Yi).
YUE Qian
Institute of Applied Ecology, Chinese Academy of Sciences
Tel: 86-24-83970317
E-mail: yueqian@iae.ac.cn
Web: http://english.iae.cas.cn



